ModisDownload: an R function to download, mosaic, and reproject the MODIS images
[ModisDownload is included in the rts package!]
MODIS instruments including two satellites (Terra and Aqua) are viewing the entire earth surface every 1 to 2 days, acquiring 36 spectral bands. These data provide insight on global processes occurring on the land, ocean and atmosphere. The images ranging in wavelength from 0.4 µm to 14.4 µm, at spatial resolutions of 250, 500, and 1000 m. You can find an overview and more details on image specifications here.
This page introduces you an implemented function in R that can be used to download, mosaic and reproject of MODIS images from Land processes distributed active archive center (LP DAAC). Thanks Tomislav Hengl, as his script made the core of the ModisDownload function.
What does ModisDownload offer?
‘ModisDownload’ downloads a series of images in a specific date or a date range and in specified tile(s). It also uses MODIS Reproject Tool (MRT) software to mosaic the downloaded images, in case of selecting more than one tile, and reproject them to specified coordinate system. As the format of the source images in LP DAAC is HDF, this tool also can convert them into other formats (i.e. Geotif, hdr).
Instructions to use ModisDownload:
In order to use ModisDownload, you first need:
-
To be able to download the data, you need to register on "https://urs.earthdata.nasa.gov/" and get a username and password. To pass the authentication by the website, you need to set the username and password on the machine in your R session (only first time) using the setNASAauth function in the rts package.
- Download and install MRT on your machine. Keep in mind, after installing MRT, you need to restart your machine. (If you do not need to mosaic, convert, and reproject the downloaded images, you do not need MRT!).
- ModisDownload is included in the rts package. So, by installing "rts" on your machine, you can simply use ModisDownload.
Following, you will find more instructions and examples on how to work with functions offered by ModisDownload (you can also check the help page of the function in the rts package):
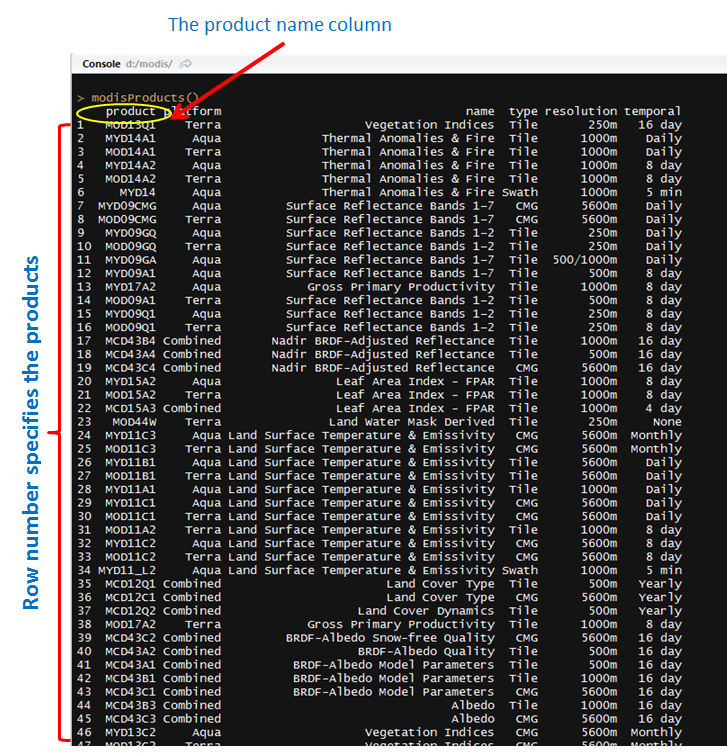
a) First specify the name or the code of the MODIS product you need. To get the product list, you can simply run the modisProducts() command. It gives you a table including the product names that can be downloaded by the functions offered here. You can either take the name of the product from the first column or the row number where the product you need is specified.
In R, by calling modisProducts() you can get the list.
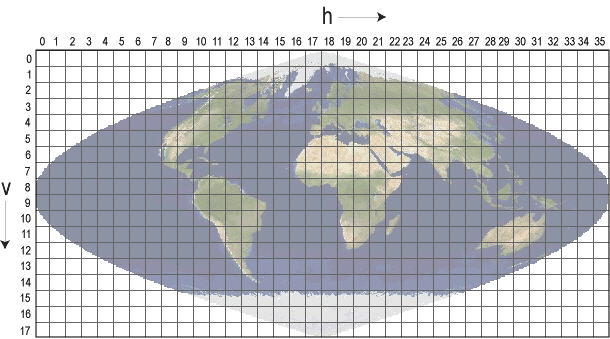
b) Specify the image blocks (tiles): you need to know the h/v position of the tiles (MODIS tiles on Google earth). For example, the h and v parameters for 4 tiles of h17v04, h17v05, h18v04, h18v05 would be: h=c(14,15) and v=c(4,5)
c) Specify a date range of image(s). For example, with dates = c(“2008.05.10”,”2008.05.31”) all available images between two specified dates will be downloaded, and with dates = “2008.05.11” the available images for only this single day will be downloaded.
d) Several functions are offered by ModisDownload including:
- modisProducts(): gives you a table including the modis products available for download from Land processes distributed active archive center (LP DAAC).
- getMODIS: can be used to download the original images from the http website from Land processes distributed active archive center (LP DAAC). The format of the images in the archive is HDF4.
- mosaicHDF: can be used to mosaic several images with HDF format using MODIS Reproject Tool (MRT) software. You need to have the MRT software installed on your machine and specify the path for the function.
- reprojectHDF: can be used to reproject an image with HDF format using MODIS Reproject Tool (MRT) software. This function transform the image into the specified projection and changes the format into Tiff.
- ModisDownload: can be used to download, mosaic and reproject the modis product within a run (as an alternative of the above three functions).
Usage of ModisDownload:
ModisDownload(x, h, v, version='005',dates, mosaic=FALSE, MRTpath, bands_subset = "", del=FALSE, proj= FALSE, UL ="", LR="", resample_type="NEAREST_NEIGHBOR", proj_type="UTM", proj_params, utm_zone,pixel_size)
Arguments
| x | modis product name or a number specifies the product row in the data.frame generated by modisProducts( ) function. |
| h and v | h/v code related to the position of selected Modis tile(s). Examples: h=15; h=c(14,15); v=c(4:7) |
| dates | logical. Only relevant if more than one tile is selected. If TRUE, it means you want to mosaic the tiles in a single image |
| MRTpath | Path to the bin folder into the installed directory of MRT software. It is needed only if you need to mosaic and/or reproject the images. Esample: MRTpath="d:/MRT/bin" |
| Bands_subset | HDF-EOS input files contain several layers of data (bands). This argument lets you select a subset of bands. You need to know the number and the order of bands, and then select your subset as it is shown in the following example. Suppose your image has 6 bands and you only need the second band. The parameter should be entered as: bands_subset="0 1 0 0 0 0" |
| delete | Logical. If you selected to mosaic the tiles into a single image, this argument defines whether the original HDF files should be deleted or not. |
| proj | Logical. TRUE means you want to reproject the images. Then MRTproj function will be called to reproject the images. You can also separately call MRTproj for each single HDF.The rest of arguments is related to MRTproj function which reproject an HDF file. |
| UL | Optional. Upper Left coordinate (x,y) in output coordinate system. Only if you want to spatially subset your images. |
| LR | Optional. Lower right coordinate(x,y). Only if you want to spatially subset your images. |
| resample_type |
Resampling kernel type (CUBIC_CONVOLUTION, NEAREST_NEIGHBOR, or
BILINEAR). The default is NEAREST_NEIGHBOR. |
| proj_type | Output projection short name. Valid values are AEA (Albers EqualArea), ER (Equirectangular), GEO (Geographic), IGH (Interrupted Goode Homolosine), HAM (Hammer), ISIN (Integerized Sinusoidal), LA (Lambert Azimuthal Equal Area), LCC (Lambert Conformal Conic), MERCAT (Mercator), MOL (Molleweide), PS (Polar Stereographic), SIN (Sinusoidal), TM (Transverse Mercator), and UTM (Universal Transverse Mercator). |
| proj_params | Output projection parameters. This quoted, floating-point list includes up to 15 projection parameters, with each value separated by white space (“p1 p2 ... p15”). If there are fewer than 15 values specified in the list, the remaining values will be set to zero.Integer values will automatically be converted to floating point. |
| datum='WGS84' | Specifies the output projection datum. The default is WGS84 |
| utm_zone | Valid only if "UTM" is selected for the proj_type. |
| pixel_size | Output pixel size. |
ModisDownload function calls reprojectHDF function when the proj argument is set to TRUE. reprojectHDF, however, can be called separately to reproject and subset of a HDF image. The usage of reprojectHDF is:
Usage of reprojectHDF:
reprojectHDF(hdfName,filename,MRTpath,UL="",LR="", resample_type="NEAREST_NEIGHBOR", proj_type="UTM", proj_params = '0 0 0 0 0 0 0 0 0 0 0 0' ,datum='WGS84', utm_zone,pixel_size)
Arguments
| hdfName | The input image (has HDF format). example: hdfName="my_image.hdf" |
| filename | The output image name. Its extension defines the format of output image and can be one of these three formats: hdf, hdr, tif. example: outname="out_image.tif" |
| MRTpath | Path to the bin folder into the installed directory of MRT software. It is needed only if you need to mosaic and/or reproject the images. Esample: MRTpath="d:/MRT/bin" |
The rest of arguments are the same as ModisDownload function.
Following examples will show you how you can use ModisDownload for different purposes:
For the first time using ModisDownload, you need to set the username and password you got from https://urs.earthdata.nasa.gov/, using setNASAauth(username='your_username',password='your_password')
# setting the working directory:
setwd('d:/modis')
library(rts)
library(raster)
library(RCurl)
# product list:
modisProducts( )
x=3 # or x="MOD14A1"
# download 4 tiles (h14v04, h14v05, h15v04, h15v05) in single date (2011.05.01)
# Following command only downloads the source HDF images, no mosaic and no projection
ModisDownload(x=x,h=c(17,18),v=c(4,5),dates='2011.05.01',mosaic=F,proj=F)
# alternatively, you can use modisHDF to download only HDF images:
modisHDF(x=x,h=c(17,18),v=c(4,5),dates='2011.05.01')
# same as the above command, but downloads all available images in 2011:
ModisDownload(x=x,h=c(17,18),v=c(4,5),dates=c('2011.01.01','2011.12.31'))
#------ if you need different version of the product, let's say 006, you can simply put version="006" in the funcion
# Downloads selected tiles, and mosaic them, but no projections:
ModisDownload(x=x,h=c(17,18),v=c(4,5),dates=c('2011.05.01','2011.05.31'),MRTpath='d:/MRT/bin',mosaic=T,proj=F)
#--- alternatively, you can first download the HDF images using getMODIS, and then mosaic them using mosaicHDF!
# Downloads selected tiles, and mosaic, reproject them in UTM_WGS84, zone 30 projection and convert all bands into Geotif format (the original HDF will be deleted!):
ModisDownload(x=x,h=c(17,18),v=c(4,5),dates=c('2011.05.01','2011.05.31'),MRTpath='d:/MRT/bin', mosaic=T,proj=T,proj_type="UTM",utm_zone=30,datum="WGS84",pixel_size=1000)
# Same as above command, but only second band out of 6 bands will be kept. (You do not need to specify proj_params when "UTM" is selected as proj_type and the zone also is specified, but for other types of projections you do).
ModisDownload(x=x,h=c(17,18),v=c(4,5),dates=c('2011.05.01','2011.05.31'),MRTpath='d:/MRT/bin',mosaic=T,proj=T, bands_subset="0 1 0 0 0 0", proj_type="UTM",proj_params="-3 0 0 0 0 0 0 0 0 0 0 0 0 0 0",utm_zone=30,datum="WGS84",pixel_size=1000)
# Same as above command, but it spatially subsets the images into the specified box (UL and LR):
ModisDownload(x=x,h=c(17,18),v=c(4,5),dates=c('2011.05.01','2011.05.31'),MRTpath='d:/MRT/bin',mosaic=T,proj=T,UL=c(-42841.0,4871530.0),LR=c(1026104,3983860), bands_subset="0 1 0 0 0 0", proj_type="UTM",proj_params="-3 0 0 0 0 0 0 0 0 0 0 0 0 0 0",utm_zone=30,datum="WGS84",pixel_size=1000)
In order to load, work, and visualize the downloaded images, the raster and rgdal packages can be used. Following, a simple example is provided that show you how to read and plot the images: You can also use rts package to create an raster time series object. Very soon, these functions will be included to rts package.
# setting the working directory:
setwd('d:/modis')
library(raster)
# A function to list the files, ended with '.tif', in the working directory
listfiles <- list.files(pattern='.tif$')
listfiles
# function to read an image or images
lai <- raster("MOD15A2_2011_05_25.Lai_1km.tif")
lai
# Multiplying to the scale factor of LAI
lai <- lai * 0.1
plot(lai)
References
- Hengl,T. Download and resampling of Modis Images, Online article.
- Huete, A., Didan, K., Miura, T., Rodriguez, E.P., Gao, X., Ferreira, L.G., (2002). Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sensing of Environment, 83: 195–213.
-
Justice CO, Vermote E, Townshend JRG, Defries R, Roy DP, Hall DK, Salomonson VV, Privette JL, Riggs G, Strahler A, Lucht W, Myneni RB, Knyazikhin Y, Running SW, Nemani RR, Wan Z, Huete AR, van Leeuwen W, Wolfe RE, Giglio L, Muller J, Lewis P, Barnsley MJ. (1998). The Moderate Resolution Imaging Spectroradiometer (MODIS): land remote sensing for global change research. IEEE T Geosci Remote, 36:1228-1249
- Land Processes DAAC. (2008). MODIS Reprojection Tool User’s Manual. USGS Earth Resources Observation and Science (EROS) Center
